-
Categories
-
Pharmaceutical Intermediates
-
Active Pharmaceutical Ingredients
-
Food Additives
- Industrial Coatings
- Agrochemicals
- Dyes and Pigments
- Surfactant
- Flavors and Fragrances
- Chemical Reagents
- Catalyst and Auxiliary
- Natural Products
- Inorganic Chemistry
-
Organic Chemistry
-
Biochemical Engineering
- Analytical Chemistry
-
Cosmetic Ingredient
- Water Treatment Chemical
-
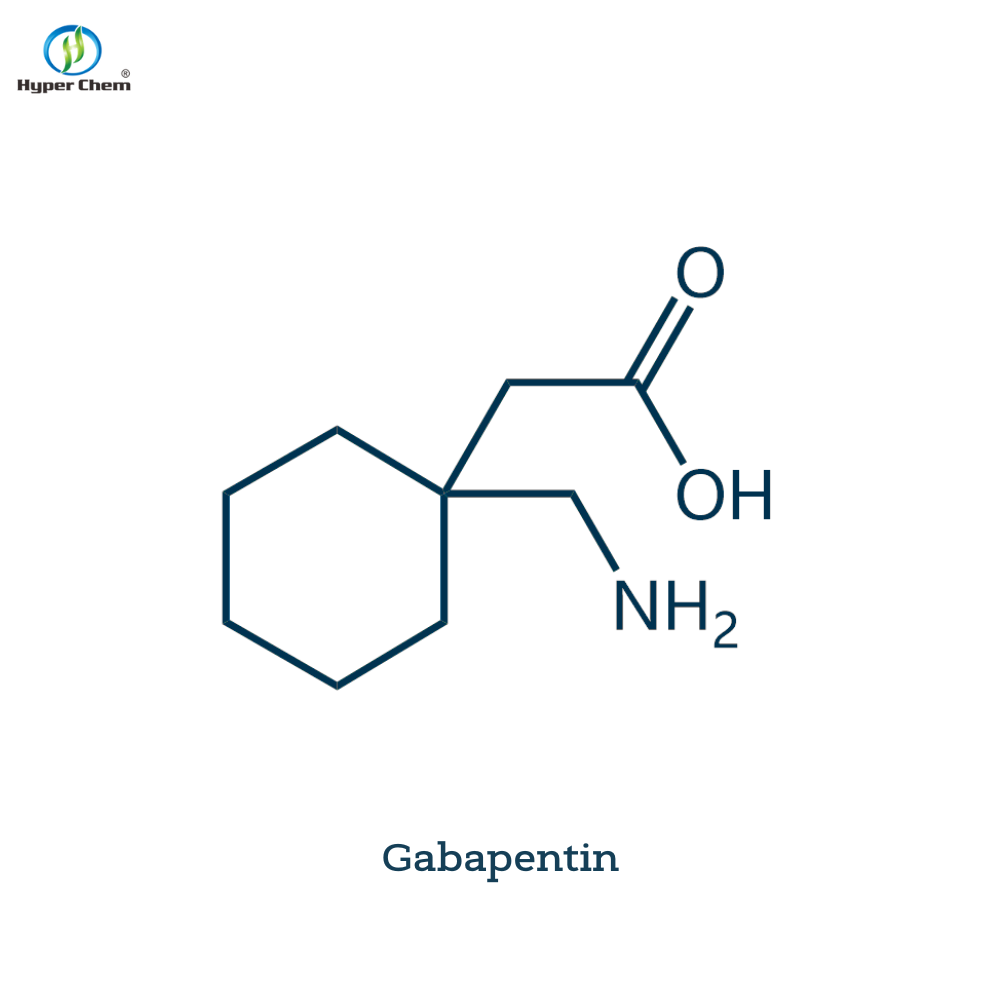
Pharmaceutical Intermediates
Promotion
ECHEMI Mall
Wholesale
Weekly Price
Exhibition
News
-
Trade Service
Editor | The brain is a major energy consuming organ in organs-it only accounts for 2% of body weight, but consumes 20% of the body's energy.
Above the high basal metabolic rate, different cognitive processes will activate the neural activity of different brain regions, thereby further increasing the local metabolic demand.
In order to ensure the smooth progress of one after another in the brain, metabolic substrates such as glucose and oxygen need to be replenished to every corner of the brain in time, and metabolic wastes need to be cleared in time.
It is the blood vessel network of about 800 kilometers in our brain that undertakes this difficult task.
So, how does the vascular network meet the metabolic needs of the brain that are constantly changing in time and space? Locally activated neural activity is often accompanied by an increase in local blood flow (neuro-vascular coupling), but the modulation amplitude of blood flow in a healthy brain is generally less than 30%, and autoradiography and positron emission tomography show different results The basal metabolic rate of the brain area is several times different.
Therefore, a considerable part of the differences in the metabolism of brain regions may be reflected in the static structure of the local blood vessels.
On the other hand, many functional imaging and magnetic resonance imaging (MRI) are based on the nerve-vascular coupling mechanism, and the signal generation is also closely linked to the vascular structure.
The measurement and understanding of cerebrovascular structure is not only an important and basic physiological problem, but also provides basic information for further quantitative analysis of signals from MRI and other technologies, and even tissue engineering.
March 2, 2021, the David Kleinfeld laboratory of the Department of Physics and the Department of Neurobiology, University of California, San Diego, and the Howard Hughes Medical Institute Janelia Research Park MouseLight team (the first author is Ji Xiang, Department of Physics, University of California, San Diego) In the Neuron article "Brain microvasculature has a common topology with local differences in geometry that match metabolic load", the microvascular network of the whole brain of mice was measured, reconstructed and analyzed.
The researchers used the two-photon microscope to image the blood vessels infused with fluorescent dyes with sub-micron resolution, and developed a set of calculation tools for image stitching, segmentation, vessel skeleton extraction, radius estimation and calibration, and network structure correction.
Finally, the image data of about 20,000 GB (per brain) is transformed into a digital blood vessel network that records the coordinates, radii, and connections of about 6 million microvessel fragments in the whole brain.
Based on this microvascular "architectural drawing", the static three-dimensional structure of the network can be completely reconstructed.
Starting from this, the researchers analyzed the similarities and differences of microvascular networks in different brain regions.
On the one hand, the researchers found that the topological structures of blood vessel networks in different brain regions are highly similar.
This may correspond to a similar morphological development process, and also suggests a similar network structure stability.
Numerical experiments have shown that in different brain regions, when about 44% of blood vessel fragments are randomly removed, the microvascular network suddenly collapses.
This threshold indicates the structural limit of the cerebrovascular network to maintain function under injury and blockage, which may be related to the development of cerebral small vessel disease and the recovery of reperfusion after stroke.
On the other hand, the researchers found that the linear density of blood vessels in different brain regions is about 3 times different.
In order to understand the relationship between blood vessel density and metabolism, the researchers analyzed the distribution of the distance between blood vessels and brain parenchymal tissues, and found that the seemingly complex network of microvessels is relatively evenly interwoven in the space at the scale of hundreds of microns, and the tissues and The maximum distance of blood vessels can be closely linked to the line density of blood vessels through a simple geometric scaling relationship.
On this basis, the researchers combined theoretical derivation and numerical calculations to propose a biophysical model linking cerebral blood vessel density and glucose metabolism rate, and further predicted by comparing the measured local density with the resting glucose metabolism rate in the literature.
Different brain regions have similar tissues-the maximum oxygen concentration of blood vessels is different.
More generally, the model and data show that the linear density of the microvascular network matches the resting metabolic rate, so that the minimum (large) concentration of the metabolic substrate (product) transported by diffusion in different brain regions in the tissue is approximately the same.
In addition, the researchers also found that a considerable part of the microvascular network has a significant orientation preference (anisotropy).
In areas with a high degree of anisotropy, the signals of some MRI techniques may significantly depend on the measurement angle, suggesting that the anatomical structure of brain capillaries may be a factor that cannot be ignored in quantitative interpretation and comparison of high-resolution MRI measurements.
All in all, this article reports the sub-micron resolution reconstruction of the whole rat brain microvascular network, revealing the similarity of the microvascular network topology in the whole brain and the geometric structure difference matching the metabolic rate, which is a further quantitative simulation.
The regulation of cerebral blood flow, material transport and even the design of artificial blood vessel networks provide basic information.
Reprinting instructions [Non-original articles] The copyright of this article belongs to the author of the article.
Personal forwarding and sharing are welcome.
Reprinting is prohibited without permission.
The author has all legal rights and offenders must be investigated.
Original link: https://doi.
org/10.
1016/j.
neuron.
2021.
02.
006 Platemaker: Qi Jiang
Above the high basal metabolic rate, different cognitive processes will activate the neural activity of different brain regions, thereby further increasing the local metabolic demand.
In order to ensure the smooth progress of one after another in the brain, metabolic substrates such as glucose and oxygen need to be replenished to every corner of the brain in time, and metabolic wastes need to be cleared in time.
It is the blood vessel network of about 800 kilometers in our brain that undertakes this difficult task.
So, how does the vascular network meet the metabolic needs of the brain that are constantly changing in time and space? Locally activated neural activity is often accompanied by an increase in local blood flow (neuro-vascular coupling), but the modulation amplitude of blood flow in a healthy brain is generally less than 30%, and autoradiography and positron emission tomography show different results The basal metabolic rate of the brain area is several times different.
Therefore, a considerable part of the differences in the metabolism of brain regions may be reflected in the static structure of the local blood vessels.
On the other hand, many functional imaging and magnetic resonance imaging (MRI) are based on the nerve-vascular coupling mechanism, and the signal generation is also closely linked to the vascular structure.
The measurement and understanding of cerebrovascular structure is not only an important and basic physiological problem, but also provides basic information for further quantitative analysis of signals from MRI and other technologies, and even tissue engineering.
March 2, 2021, the David Kleinfeld laboratory of the Department of Physics and the Department of Neurobiology, University of California, San Diego, and the Howard Hughes Medical Institute Janelia Research Park MouseLight team (the first author is Ji Xiang, Department of Physics, University of California, San Diego) In the Neuron article "Brain microvasculature has a common topology with local differences in geometry that match metabolic load", the microvascular network of the whole brain of mice was measured, reconstructed and analyzed.
The researchers used the two-photon microscope to image the blood vessels infused with fluorescent dyes with sub-micron resolution, and developed a set of calculation tools for image stitching, segmentation, vessel skeleton extraction, radius estimation and calibration, and network structure correction.
Finally, the image data of about 20,000 GB (per brain) is transformed into a digital blood vessel network that records the coordinates, radii, and connections of about 6 million microvessel fragments in the whole brain.
Based on this microvascular "architectural drawing", the static three-dimensional structure of the network can be completely reconstructed.
Starting from this, the researchers analyzed the similarities and differences of microvascular networks in different brain regions.
On the one hand, the researchers found that the topological structures of blood vessel networks in different brain regions are highly similar.
This may correspond to a similar morphological development process, and also suggests a similar network structure stability.
Numerical experiments have shown that in different brain regions, when about 44% of blood vessel fragments are randomly removed, the microvascular network suddenly collapses.
This threshold indicates the structural limit of the cerebrovascular network to maintain function under injury and blockage, which may be related to the development of cerebral small vessel disease and the recovery of reperfusion after stroke.
On the other hand, the researchers found that the linear density of blood vessels in different brain regions is about 3 times different.
In order to understand the relationship between blood vessel density and metabolism, the researchers analyzed the distribution of the distance between blood vessels and brain parenchymal tissues, and found that the seemingly complex network of microvessels is relatively evenly interwoven in the space at the scale of hundreds of microns, and the tissues and The maximum distance of blood vessels can be closely linked to the line density of blood vessels through a simple geometric scaling relationship.
On this basis, the researchers combined theoretical derivation and numerical calculations to propose a biophysical model linking cerebral blood vessel density and glucose metabolism rate, and further predicted by comparing the measured local density with the resting glucose metabolism rate in the literature.
Different brain regions have similar tissues-the maximum oxygen concentration of blood vessels is different.
More generally, the model and data show that the linear density of the microvascular network matches the resting metabolic rate, so that the minimum (large) concentration of the metabolic substrate (product) transported by diffusion in different brain regions in the tissue is approximately the same.
In addition, the researchers also found that a considerable part of the microvascular network has a significant orientation preference (anisotropy).
In areas with a high degree of anisotropy, the signals of some MRI techniques may significantly depend on the measurement angle, suggesting that the anatomical structure of brain capillaries may be a factor that cannot be ignored in quantitative interpretation and comparison of high-resolution MRI measurements.
All in all, this article reports the sub-micron resolution reconstruction of the whole rat brain microvascular network, revealing the similarity of the microvascular network topology in the whole brain and the geometric structure difference matching the metabolic rate, which is a further quantitative simulation.
The regulation of cerebral blood flow, material transport and even the design of artificial blood vessel networks provide basic information.
Reprinting instructions [Non-original articles] The copyright of this article belongs to the author of the article.
Personal forwarding and sharing are welcome.
Reprinting is prohibited without permission.
The author has all legal rights and offenders must be investigated.
Original link: https://doi.
org/10.
1016/j.
neuron.
2021.
02.
006 Platemaker: Qi Jiang