-
Categories
-
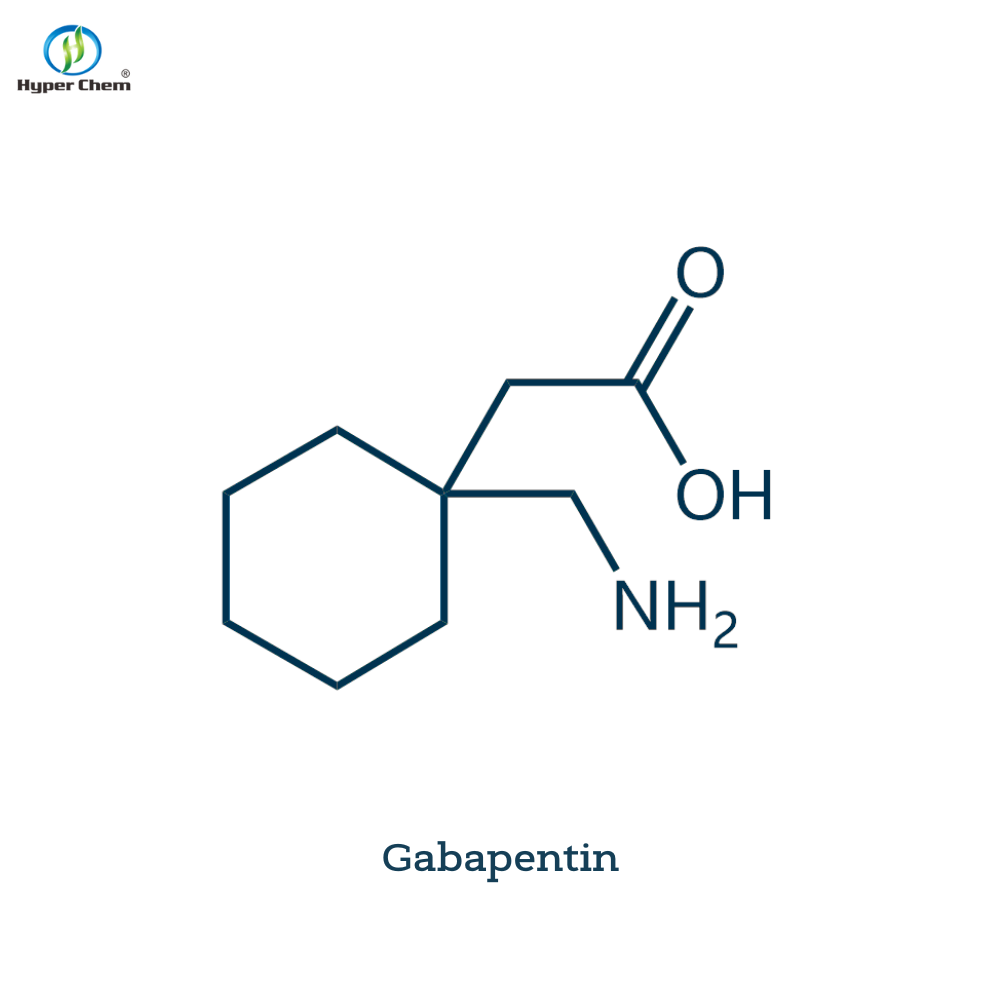
Pharmaceutical Intermediates
-
Active Pharmaceutical Ingredients
-
Food Additives
- Industrial Coatings
- Agrochemicals
- Dyes and Pigments
- Surfactant
- Flavors and Fragrances
- Chemical Reagents
- Catalyst and Auxiliary
- Natural Products
- Inorganic Chemistry
-
Organic Chemistry
-
Biochemical Engineering
- Analytical Chemistry
-
Cosmetic Ingredient
- Water Treatment Chemical
-
Pharmaceutical Intermediates
Promotion
ECHEMI Mall
Wholesale
Weekly Price
Exhibition
News
-
Trade Service
Written | Yan Xiao From an anatomical point of view, the brain can be subdivided into a number of specific areas, including the neocortex (neocortex)
.
The cerebral cortex is the center of advanced cognition and the most expanded and diversified area in the brain during human evolution
.
Early brain divisions and cortical divisions were guided by morphogenetic gradients [1-2], but as the development process unfolds, how these early patterns produce more detailed and discrete spatial differences is currently unclear.
【3】
.
The development process of the cerebral cortex has been studied for more than a century.
Historically, scientists have explored by observing only one cell type at a time, studying a small number of genes, and then gradually splicing the entire developmental event
.
But we must realize that the brain does not only produce one cell type at the same time, but hundreds of cell types develop together, which is as beautiful and complicated as a symphony
.
With the emergence and development of single-cell and spatial transcriptomics, combined with big data analysis, we have been able to explore the laws hidden in the symphony of neurodevelopment
.
On October 6, 2021, the team of Arnold R.
Kriegstein from the University of California published an online research paper entitled An atlas of cortical arealization identifies dynamic molecular signatures in Nature
.
The study used single-cell sequencing to study 10 main brain regions and 6 neocortical regions in the stages of neurodevelopment and early glial production, revealing the molecular maps of the longitudinal development of different cells in different cortical regions
.
Drawing the human brain development map In order to depict the cell diversity of different brain regions and cortical regions during brain development, the author collected brain tissues in the second trimester (3-6 months of pregnancy, peak neurodevelopment), and then divided them (brain The subdivided regions are called "regions", and the cortical subdivided regions are called "areas") and single-cell transcription sequencing (Figure 1)
.
The author obtained 10 brain regions (mainly forebrain, midbrain and hindbrain) samples and 6 neocortical region samples (prefrontal cortex (PFC), motor, somatosensory, parietal, temporal and primary visual (V1) from 13 individuals ) Cortex), and finally obtained 698,820 high-quality single-cell data
.
Through UMPA (uniform manifold approximation and projection, new dimensionality reduction technology for data visualization and exploration) analysis, the author found the expected cell groups (including exciting neurons, intermediate progenitor cells (IPCs), radial glia, etc.
)
.
The data shows that in the whole brain, the cell type is the main factor that produces regional differentiation and isolation
.
Analysis of region-specific genes showed that some region-specific genes exist in multiple cell types in the same region, indicating that certain regionally expressed gene characteristics are highly permeable in cell types
.
Figure 1.
Schematic diagram of sequencing sample collection.
Cell types in the neocortex.
Studies have shown that the neocortex includes dozens of functional areas dedicated to cognitive processes [4]
.
Neurons in V1 and PFC are completely different after birth [5], while other cell types do not show obvious regional-specific differences
.
In order to further expand the existing research, the authors sequenced and analyzed single cells from specific cortical areas, and obtained 387,141 high-quality single-cell data
.
Through analysis, the authors found that the desired cell types, including the Cajal-Retzius neurons, dividing cells, excitatory neurons and so on
.
Subsequently, hierarchical clustering by cell type resulted in 138 neocortical cell populations, of which 104 cell populations were composed of cells from multiple cortical regions
.
Dynamic regional gene characteristics In order to explore the cell regional differences in the development of the neocortex, the authors searched for the differentially expressed genes in each cell type in the excitatory lineages (radial glial (RG), IPCs and excitatory neurons) in different regions of the cortex At the same time, the reliability of cortical region division is evaluated by detecting the expression of known region-specific genes
.
The author constructed a constellation diagram to explore the relationship between cell types in different cortical regions: RG nodes are mainly connected to each other between the same cell type; there are interconnections between IPC and excitatory neurons; there is no connection between PFC and V1 cell type nodes , Indicating that the two gene expression patterns are mutually exclusive
.
In each set of regional marker genes, the authors identified genes encoding transcription factors, which are abundantly enriched in cells in specific regions
.
These include some transcription factors with known functions in the process of regionalization, such as NR2F1 and BCL11A, both of which are related to neurodevelopmental diseases [6]
.
The author also found some transcription factors that are not related to cortical regionalization: In V1, including NF1A, NF1B and NF1X, they are important regulators of brain development and are related to macrocephaly and cognitive impairment [7]; ZBTB18, brain expansion Driving factors are related to neuron differentiation and cortical migration; in PFC, including HMGB2 and HMGB3, they are differentially expressed in neural stem cells at different stages of development, and are key regulators of neural differentiation, but they are regionalized in the cortex The function of the process has not been studied and reported
.
Validation of candidate markers by in situ hybridization The above single-cell data revealed the diversity and transcription profile of cell types in six different regions of the cortex during human brain development
.
Next, the author selected candidate marker genes of excitatory neuron clusters for verification, and quantified 20 samples (from 4 cortical regions) using single-molecule fluorescent in situ hybridization (smFISH).
The expression of 31 RNA transcripts (Figure 2)
.
Consistent with previous reports, the neurogenes SATB2 and BCL11B show regional dynamic expression: they are co-expressed in the frontal region, but mutually exclusive in the occipital region
.
By analyzing all regions, the author found new candidate genes for sub-cell population markers: NEFL, SERPINI1 and NR4A1
.
The expression levels of these three genes in PFC, somatosensory, temporal and V1 cortical cells are basically equal, but their relative spatial positions have changed dramatically: NEFL, SERPINI1 and NR4A1 are co-expressed in PFC, but are mutually exclusive in other regions; In the somatosensory cortex, these marker genes are mainly expressed in the upper molecular layer
.
Figure 2.
Automated spatial RNA transcription detection process.
In summary, this study provides a detailed understanding of the gene expression characteristics of different cell types in the neocortex region
.
The authors found that: (1) In the main brain structure, regional features are very common in different cell types; (2) Regional features in the neocortex are very special and limited to a single cell type; (3) In addition to cell type features In addition, the developmental stage of the cell (ie, pregnancy week) is a powerful determinant of the combination of gene expression characteristics
.
These findings indicate that the dynamic changes of region-specific gene expression characteristics are very fast and cell-type-specific (Figure 3).
This does not seem to be consistent with previous theories.
In previous cognitions, gene expression patterns are usually It is believed that once established, it will continue to exist
.
By mapping gene expression during brain development, researchers have a better understanding of how the cerebral cortex is formed, which helps to explore how the cerebral cortex is affected in neurodevelopmental diseases
.
Figure 3.
Cortex regionalization pattern map during development.
Original link: https://doi.
org/10.
1038/s41586-021-03910-8 Plate maker: Eleven references [1] Cadwell, CR, Bhaduri, A.
, Mostajo-Radji, et al.
Development and arealization of the cerebral cortex.
Neuron 103, 980-1004 (2019).
[2] O'Leary, DD, Chou, SJ & Sahara, S.
Area patterning of the mammalian cortex.
Neuron 56, 252-269 (2007).
[3] Tasic, B.
et al.
Shared and distinct transcriptomic cell types across neocortical areas.
Nature 563, 72–78 (2018).
[4] Rakic, P.
Evolution of the neocortex : a perspective from developmental biology.
Nat.
Rev.
Neurosci.
10, 724-735 (2009).
[5] Nowakowski, TJ et al.
Spatiotemporal gene expression trajectories reveal developmental hierarchies of the human cortex.
Science 358, 1318-1323 ( 2017).
[6] Bergen, V.
, Lange, M.
, Peidli, S.
, et al.
Generalizing RNA velocity to transient cell states through dynamical modeling.
Nat.
Biotechnol.
38, 1408-1414 (2020).
[7] Butler, A.
, Hoffman, P.
, Smibert, P.
, et al.
Integrating single-cell transcriptomic data across different conditions, technologies, and species.
Nat.
Biotechnol 36, 411-420 (2018).
Reprinting instructions [Original Articles] BioArt original articles, personal reposting and sharing are welcome, reprinting is prohibited without permission, the copyrights of all published works are owned by BioArt
.
BioArt reserves all statutory rights and offenders must be investigated
.
.
The cerebral cortex is the center of advanced cognition and the most expanded and diversified area in the brain during human evolution
.
Early brain divisions and cortical divisions were guided by morphogenetic gradients [1-2], but as the development process unfolds, how these early patterns produce more detailed and discrete spatial differences is currently unclear.
【3】
.
The development process of the cerebral cortex has been studied for more than a century.
Historically, scientists have explored by observing only one cell type at a time, studying a small number of genes, and then gradually splicing the entire developmental event
.
But we must realize that the brain does not only produce one cell type at the same time, but hundreds of cell types develop together, which is as beautiful and complicated as a symphony
.
With the emergence and development of single-cell and spatial transcriptomics, combined with big data analysis, we have been able to explore the laws hidden in the symphony of neurodevelopment
.
On October 6, 2021, the team of Arnold R.
Kriegstein from the University of California published an online research paper entitled An atlas of cortical arealization identifies dynamic molecular signatures in Nature
.
The study used single-cell sequencing to study 10 main brain regions and 6 neocortical regions in the stages of neurodevelopment and early glial production, revealing the molecular maps of the longitudinal development of different cells in different cortical regions
.
Drawing the human brain development map In order to depict the cell diversity of different brain regions and cortical regions during brain development, the author collected brain tissues in the second trimester (3-6 months of pregnancy, peak neurodevelopment), and then divided them (brain The subdivided regions are called "regions", and the cortical subdivided regions are called "areas") and single-cell transcription sequencing (Figure 1)
.
The author obtained 10 brain regions (mainly forebrain, midbrain and hindbrain) samples and 6 neocortical region samples (prefrontal cortex (PFC), motor, somatosensory, parietal, temporal and primary visual (V1) from 13 individuals ) Cortex), and finally obtained 698,820 high-quality single-cell data
.
Through UMPA (uniform manifold approximation and projection, new dimensionality reduction technology for data visualization and exploration) analysis, the author found the expected cell groups (including exciting neurons, intermediate progenitor cells (IPCs), radial glia, etc.
)
.
The data shows that in the whole brain, the cell type is the main factor that produces regional differentiation and isolation
.
Analysis of region-specific genes showed that some region-specific genes exist in multiple cell types in the same region, indicating that certain regionally expressed gene characteristics are highly permeable in cell types
.
Figure 1.
Schematic diagram of sequencing sample collection.
Cell types in the neocortex.
Studies have shown that the neocortex includes dozens of functional areas dedicated to cognitive processes [4]
.
Neurons in V1 and PFC are completely different after birth [5], while other cell types do not show obvious regional-specific differences
.
In order to further expand the existing research, the authors sequenced and analyzed single cells from specific cortical areas, and obtained 387,141 high-quality single-cell data
.
Through analysis, the authors found that the desired cell types, including the Cajal-Retzius neurons, dividing cells, excitatory neurons and so on
.
Subsequently, hierarchical clustering by cell type resulted in 138 neocortical cell populations, of which 104 cell populations were composed of cells from multiple cortical regions
.
Dynamic regional gene characteristics In order to explore the cell regional differences in the development of the neocortex, the authors searched for the differentially expressed genes in each cell type in the excitatory lineages (radial glial (RG), IPCs and excitatory neurons) in different regions of the cortex At the same time, the reliability of cortical region division is evaluated by detecting the expression of known region-specific genes
.
The author constructed a constellation diagram to explore the relationship between cell types in different cortical regions: RG nodes are mainly connected to each other between the same cell type; there are interconnections between IPC and excitatory neurons; there is no connection between PFC and V1 cell type nodes , Indicating that the two gene expression patterns are mutually exclusive
.
In each set of regional marker genes, the authors identified genes encoding transcription factors, which are abundantly enriched in cells in specific regions
.
These include some transcription factors with known functions in the process of regionalization, such as NR2F1 and BCL11A, both of which are related to neurodevelopmental diseases [6]
.
The author also found some transcription factors that are not related to cortical regionalization: In V1, including NF1A, NF1B and NF1X, they are important regulators of brain development and are related to macrocephaly and cognitive impairment [7]; ZBTB18, brain expansion Driving factors are related to neuron differentiation and cortical migration; in PFC, including HMGB2 and HMGB3, they are differentially expressed in neural stem cells at different stages of development, and are key regulators of neural differentiation, but they are regionalized in the cortex The function of the process has not been studied and reported
.
Validation of candidate markers by in situ hybridization The above single-cell data revealed the diversity and transcription profile of cell types in six different regions of the cortex during human brain development
.
Next, the author selected candidate marker genes of excitatory neuron clusters for verification, and quantified 20 samples (from 4 cortical regions) using single-molecule fluorescent in situ hybridization (smFISH).
The expression of 31 RNA transcripts (Figure 2)
.
Consistent with previous reports, the neurogenes SATB2 and BCL11B show regional dynamic expression: they are co-expressed in the frontal region, but mutually exclusive in the occipital region
.
By analyzing all regions, the author found new candidate genes for sub-cell population markers: NEFL, SERPINI1 and NR4A1
.
The expression levels of these three genes in PFC, somatosensory, temporal and V1 cortical cells are basically equal, but their relative spatial positions have changed dramatically: NEFL, SERPINI1 and NR4A1 are co-expressed in PFC, but are mutually exclusive in other regions; In the somatosensory cortex, these marker genes are mainly expressed in the upper molecular layer
.
Figure 2.
Automated spatial RNA transcription detection process.
In summary, this study provides a detailed understanding of the gene expression characteristics of different cell types in the neocortex region
.
The authors found that: (1) In the main brain structure, regional features are very common in different cell types; (2) Regional features in the neocortex are very special and limited to a single cell type; (3) In addition to cell type features In addition, the developmental stage of the cell (ie, pregnancy week) is a powerful determinant of the combination of gene expression characteristics
.
These findings indicate that the dynamic changes of region-specific gene expression characteristics are very fast and cell-type-specific (Figure 3).
This does not seem to be consistent with previous theories.
In previous cognitions, gene expression patterns are usually It is believed that once established, it will continue to exist
.
By mapping gene expression during brain development, researchers have a better understanding of how the cerebral cortex is formed, which helps to explore how the cerebral cortex is affected in neurodevelopmental diseases
.
Figure 3.
Cortex regionalization pattern map during development.
Original link: https://doi.
org/10.
1038/s41586-021-03910-8 Plate maker: Eleven references [1] Cadwell, CR, Bhaduri, A.
, Mostajo-Radji, et al.
Development and arealization of the cerebral cortex.
Neuron 103, 980-1004 (2019).
[2] O'Leary, DD, Chou, SJ & Sahara, S.
Area patterning of the mammalian cortex.
Neuron 56, 252-269 (2007).
[3] Tasic, B.
et al.
Shared and distinct transcriptomic cell types across neocortical areas.
Nature 563, 72–78 (2018).
[4] Rakic, P.
Evolution of the neocortex : a perspective from developmental biology.
Nat.
Rev.
Neurosci.
10, 724-735 (2009).
[5] Nowakowski, TJ et al.
Spatiotemporal gene expression trajectories reveal developmental hierarchies of the human cortex.
Science 358, 1318-1323 ( 2017).
[6] Bergen, V.
, Lange, M.
, Peidli, S.
, et al.
Generalizing RNA velocity to transient cell states through dynamical modeling.
Nat.
Biotechnol.
38, 1408-1414 (2020).
[7] Butler, A.
, Hoffman, P.
, Smibert, P.
, et al.
Integrating single-cell transcriptomic data across different conditions, technologies, and species.
Nat.
Biotechnol 36, 411-420 (2018).
Reprinting instructions [Original Articles] BioArt original articles, personal reposting and sharing are welcome, reprinting is prohibited without permission, the copyrights of all published works are owned by BioArt
.
BioArt reserves all statutory rights and offenders must be investigated
.