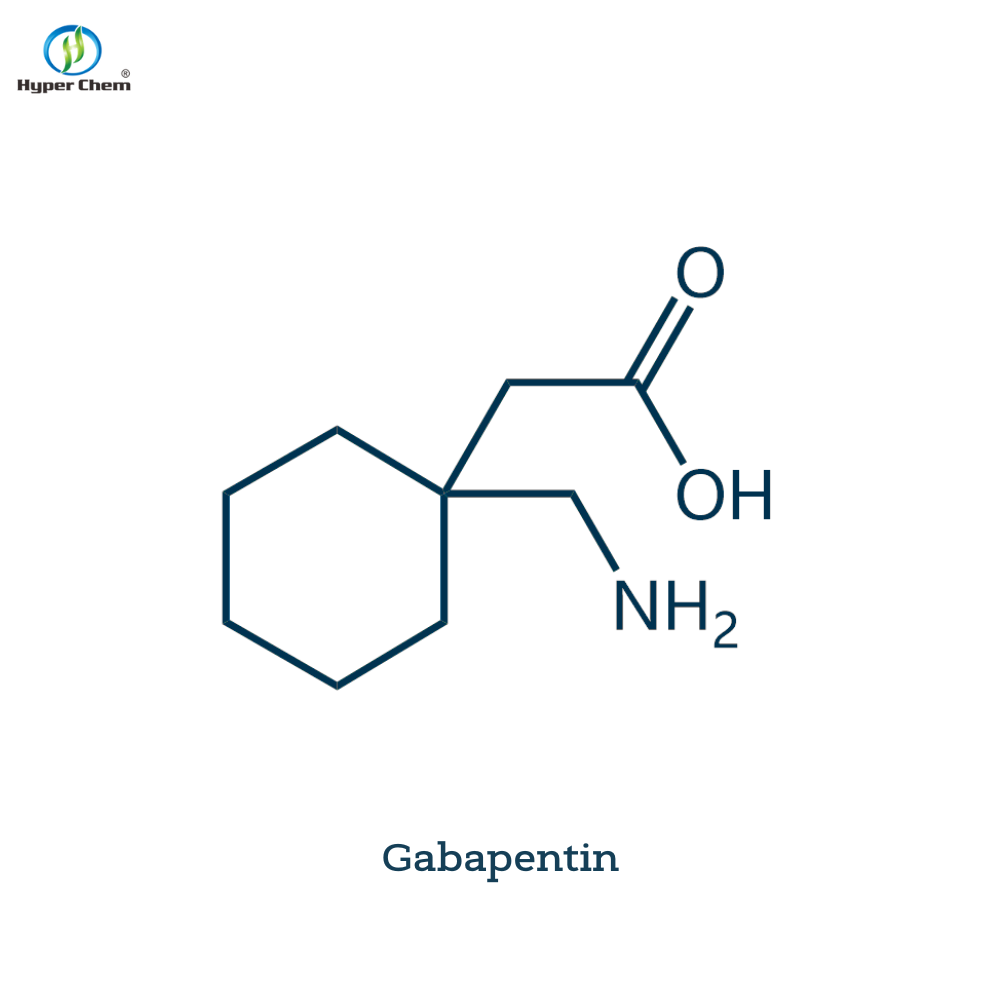
-
Categories
-
Pharmaceutical Intermediates
-
Active Pharmaceutical Ingredients
-
Food Additives
- Industrial Coatings
- Agrochemicals
- Dyes and Pigments
- Surfactant
- Flavors and Fragrances
- Chemical Reagents
- Catalyst and Auxiliary
- Natural Products
- Inorganic Chemistry
-
Organic Chemistry
-
Biochemical Engineering
- Analytical Chemistry
-
Cosmetic Ingredient
- Water Treatment Chemical
-
Pharmaceutical Intermediates
Promotion
ECHEMI Mall
Wholesale
Weekly Price
Exhibition
News
-
Trade Service
Hepatocellular carcinoma (HCC) is one of the most common malignancies and is ranked as the fourth leading cause
of cancer-related death worldwide.
Microvascular invasion (MVI), i.
e.
, HCC emboli with micrometastases within hepatic vessels, is considered a key determinant of
surgical resection and recurrence after liver transplantation.
Therefore, an accurate preoperative assessment of MVI has important clinical implications
for selecting appropriate surgical interventions.
At this stage, medical images, including CT and MR images, have been extracted to predict the MVI of HCC based on morphological characteristics; However, qualitative analysis is limited
by subjective interpretation differences.
As an MRI to assess tissue dynamics, diffusion-weighted imaging (DWI) is a promising method
for accurately predicting MVI.
Its main advantages are that there is no need to use a contrast agent and no ionizing radiation
is involved.
Multib-valued DWIs with incoherent motion within voxels (IVIM) can distinguish true molecular diffusion and microcapillary perfusion using a biexponential model, which may determine tissue perfusion and diffusion more accurately than apparent diffusion
coefficients (ADCs) calculated using a single-index model.
At present, the accuracy of IVIM-DWI in clinical application is mainly based on the quantitative description
of IVIM parameters.
However, IVIM parameters derived from the biexponential model are very sensitive to motion and artifacts in DWI[, which greatly reduces the performance
of IVIM-DWI in tumor characterization.
Developments in machine learning and artificial intelligence have added a new range
to CT and MR imaging analysis.
A previous study used a deep convolutional network to clarify the relationship between multiple b-values in DWI, and deep learning feature representations learned from multiple b-value images were used to describe cancer lesions
.
In addition, a recently published study that uses a deep convolutional network to extract the depth features of multiple b-value images in DWI can better predict the MVI
of HCC.
To our knowledge, previous studies have proposed an attention-based deep learning model to process IVIM data to improve the performance of tumor characterization, but did not take into account any clinical information and statistical analysis
.
Recently, a study published in the journal European Radiology developed and evaluated a IVIM-DWI-based preoperative MVI deep learning model to achieve accurate prediction of HCC, better characterize lesions in clinical practice to achieve prognostic prediction
.
This retrospective study included 114 patients
with pathologically confirmed HCC from December 2014 to August 2021.
All patients underwent preoperative MRI of IVIM sequences with 9 b-values
.
First, 9 b-value images are superimposed on the channel dimension to obtain a b-value volume with a shape of 32×32×9 dimensions
.
Second, the image resampling method is used for data amplification to generate more samples for training
.
Finally, the depth features
of predicting MVI in HCC are directly derived from the CNN-based b-value body.
In addition, a deep learning model based on parameter graphs and a fusion model
combining IVIM, clinical features, and deep features of IVIM parameters were constructed.
Receiver operating characteristic (ROC) curve analysis was performed to evaluate the diagnostic performance
of MVI prediction in HCC.
The depth features extracted directly from IVIM-DWI using CNNs (0.
810 (range 0.
760, 0.
829)) yielded better performance
for predicting MVI than IVIM parametric plots (0.
590 (range 0.
555, 0.
643)).
In addition, the fusion model combines the depth profile, clinical features (α-fetal protein (AFP) level and tumor size) and apparent diffusion coefficient (ADC) (0.
829 (range 0.
776, 0.
848)) with slightly improved
performance.
The Figure class activates the heat map drawn by the attention map
.
Dark red indicates a larger contribution to MVI predictions, while dark blue indicates a smaller
contribution.
a A 26-year-old male with pathologically confirmed HCC but no MVI; b A 52-year-old man with pathologically confirmed HCC for MVI
This study showed that the IVIM-based CNN model achieved high diagnostic performance
in the prediction of MVI before HCC surgery.
In addition, deep feature models that incorporate IVIM, clinical features (AFP level and tumor size), and ADC achieve better predictive performance
than IVIM-based models alone.
Therefore, the model proposed in this study can be used to predict MVI and assist clinicians in formulating precise treatment strategies
.
Original source:
Baoer Liu,Qingyuan Zeng,Jianbin Huang,et al.
IVIM using convolutional neural networks predicts microvascular invasion in HCC.
DOI:10.
1007/s00330-022-08927-9.